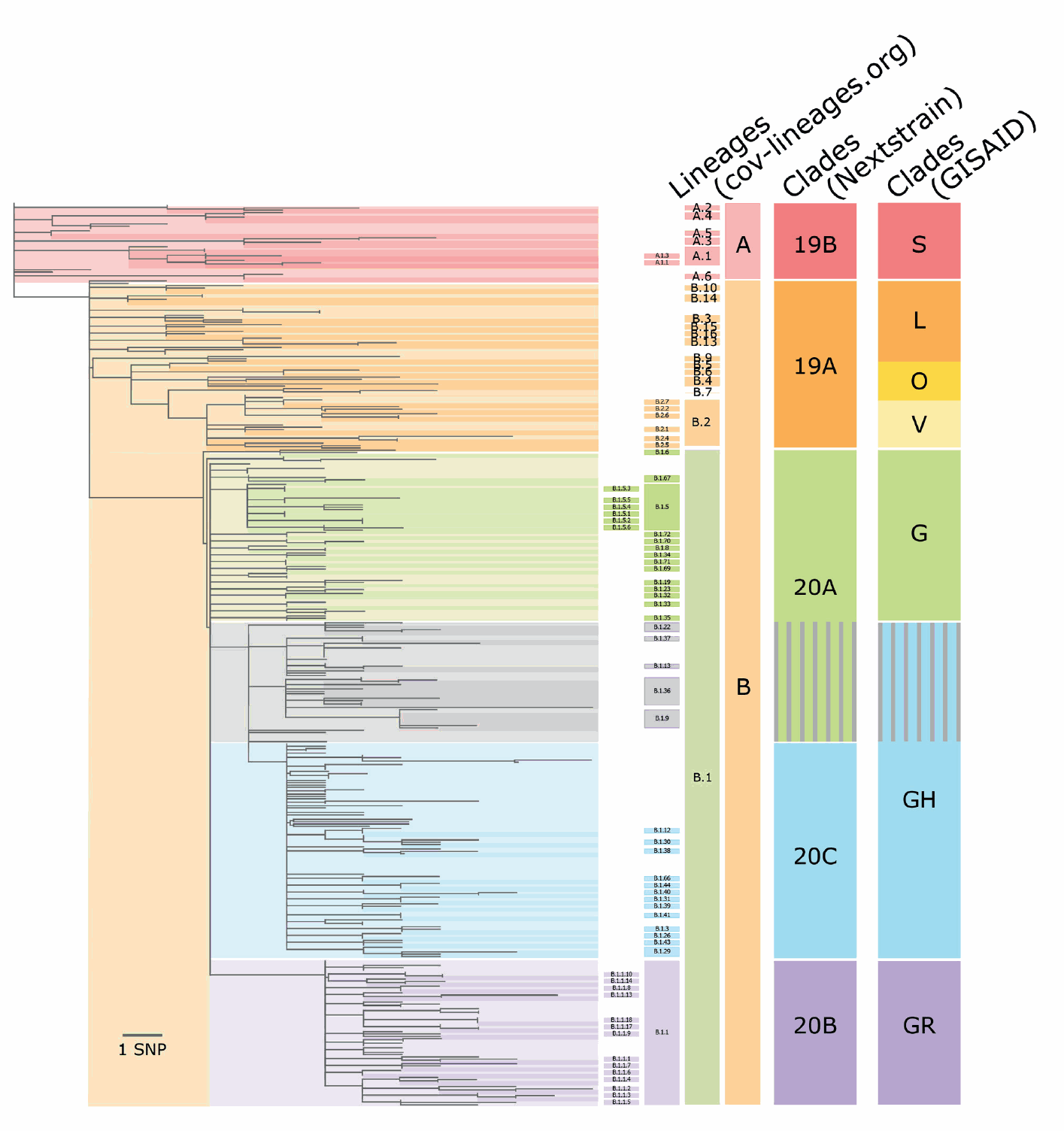
SNP: single nucleotide polymorphism.
A schematic tree is shown, representing the major topological features of the SARS-CoV-2 phylogeny. The tree is shaded to correspond to the major lineages and all Nextstrain and GISAID clades. Between Nextstrain clades 20A and 20C and GISAID clades G and GH, there are differences in the position where clades are divided (shown as grey in the main tree and lineage columns, and grey stripes in the Nextstrain and GISAID column). Here, the grey is shown as stripes in the Nextstrain and GISAID clade columns to reflect how they are categorised by each methodology. Association between the lineages and clades is approximate. The sequence data available from GISAID were curated using the Pangolin data preparation pipeline [8]. In brief, we masked out singleton substitutions identified in the pipeline and selected a set of representatives per lineage or clade. We aligned the sequences using MAFFT v7.470 [14] and estimated a maximum likelihood phylogeny using IQ-Tree v1.6.12 with 10,000 ultrafast bootstraps [15,16]. Sequence EPI_ISL_406801 (Wuhan/WH04/2020), a basal A lineage sequence, was used as an outgroup for the tree. We visualised the tree using baltic with custom python scripts and manual edits [17].

